Overview
Teaching: 30 min
Exercises: 15 minQuestions
What were the key changes when using the multiprocessing library?
How could this be implemented with
dask?How does a conversion using
daskhigh level API compare?Objectives
Use a high level description of your algorithm.
Measure the runtime of the dask implementation.
Lola observes the code she has just written. She asks her room mate if she could review it. So both of them sit down in front of the screen and go through the code again.
#!/usr/bin/env python3
import sys
import numpy as np
from multiprocessing import cpu_count, Pool
np.random.seed(2017)
def inside_circle(total_count):
x = np.float32(np.random.uniform(size=total_count))
y = np.float32(np.random.uniform(size=total_count))
radii = np.sqrt(x*x + y*y)
count = len(radii[np.where(radii<=1.0)])
return count
def estimate_pi(n_samples,n_cores):
partitions = [ ]
for i in range(n_cores):
partitions.append(int(n_samples/n_cores))
pool = Pool(processes=n_cores)
counts=pool.map(inside_circle, partitions)
total_count = sum(partitions)
return (4.0 * sum(counts) / total_count)
if __name__=='__main__':
ncores = cpu_count()
n_samples = 10000
if len(sys.argv) > 1:
n_samples = int(sys.argv[1])
partitions = [ int(n_samples/ncores) for item in range(ncores)]
sizeof = np.dtype(np.float32).itemsize
my_pi = estimate_pi(n_samples,ncores)
print("[parallel version] required memory %.3f MB" % (n_samples*sizeof*3/(1024*1024)))
print("[using %3i cores ] pi is %f from %i samples" % (ncores,my_pi,n_samples))
Lola’s office mate observes, that:
-
the libraries used are all contained in the python3 standard library (anybody else that wants to use this software only needs python3 and nothing else)
-
Lola uses implicit parallelisation by using
multiprocessing.Pool, i.e. she doesn’t need to manage her workers/threads/cores in any way -
it’s nice that Lola can reuse parts of her serial implementation
-
doing the calculation of the partitions looks quite error prone
-
larger portions of the code appear dependent on
ncores -
using
multiprocessinglimits Lola to using only one machine at a time
Lola agrees to these observations and both argue that an alternative implementation using higher level abstractions of the underlying hardware might be a good idea.
Another day, Lola discovers a library named dask (see more details
here) that not only promises high
performance, but also appears to be on par with the numpy library, so that she
has to apply only minimal changes to the code. This library can be installed on
Lola’s cluster by
$ pip3 install --user dask
She now sets out to study the documentation of dask and comes up with the
following code:
#!/usr/bin/env python3
import sys
import math
import dask.array as da
import numpy as np
np.random.seed(2017)
da.random.seed(2017)
def inside_circle(total_count, chunk_size = -1):
x = da.random.uniform(size=(total_count),
chunks=(chunk_size))
y = da.random.uniform(size=(total_count),
chunks=(chunk_size))
radii = da.sqrt(x*x + y*y)
filtered = da.where(radii <= 1.0)
indices = np.array(filtered[0])
count = len(radii[indices])
return count
def estimate_pi(total_count, chunk_size=-1):
count = inside_circle(total_count, chunk_size)
return (4.0 * count / total_count)
def main():
n_samples = 10000
if len(sys.argv) > 1:
n_samples = int(sys.argv[1])
chunksize = .1*n_samples
if len(sys.argv) > 2:
chunksize = int(sys.argv[2])
my_pi = estimate_pi(n_samples, chunksize)
sizeof = np.dtype(np.float32).itemsize
print("[parallel version] required memory %.3f MB" % (n_samples*sizeof*3/(1024*1024)))
print("[using dask lib ] pi is %f from %i samples" % (my_pi,n_samples))
if __name__=='__main__':
main()
This implementation can
now be put to work. At this point, a paradigm shift has been introduced
silently. Lola’s office mate makes her aware of this. It is a subtle change
compared to using the multiprocessing library, but it is there.
In this example, the containers for the random numbers have become smart. This
is only visible by good measure of the chunks=-1 argument to the
da.random.uniform function. A flat container used to just hold numbers in
memory wouldn’t have to be responsible for the chunking of the data. But dask
offers us a container that does so.
Behind the curtains, the dask framework connects containers (da.array here)
with functions (operator*, operator+, da.sqrt, da.where). The framework
then infers which functions can act on which data independently. From this, the
dask library can complete the program to any degree of parallelism that is
needed.
All this automation comes at a price. The dask implementation is about 2x
slower than the pure multiprocessing one. But there must be something, Lola
has gained. The answer will become evident, when we dive into more details of
the dask ecosystem as dask is HPC-ready.
$ pip3 install --user distributed bokeh
When consulting the dask.distributed
documentation, Lola
recognizes that she needs to adopt her code to work with dask.distributed.
#!/usr/bin/env python3
import sys
import math
import dask.array as da
from dask.distributed import Client
import numpy as np
np.random.seed(2017)
da.random.seed(2017)
def inside_circle(total_count, chunk_size = -1):
x = da.random.uniform(size=(total_count),
chunks=(chunk_size))
y = da.random.uniform(size=(total_count),
chunks=(chunk_size))
radii = da.sqrt(x*x + y*y)
filtered = da.where(radii <= 1.0)
indices = np.array(filtered[0])
count = len(radii[indices])
return count
def estimate_pi(total_count, chunk_size=-1):
count = inside_circle(total_count, chunk_size)
return (4.0 * count / total_count)
def main():
n_samples = 10000
if len(sys.argv) > 1:
n_samples = int(sys.argv[1])
chunksize = .1*n_samples
if len(sys.argv) > 2:
chunksize = int(sys.argv[2])
client = Client("tcp://192.168.178.25:8786")
my_pi = estimate_pi(n_samples, chunksize)
sizeof = np.dtype(np.float32).itemsize
print("[parallel version] required memory %.3f MB" % (n_samples*sizeof*3/(1024*1024)))
print("[distributed dask] pi is %f from %i samples" % (my_pi,n_samples))
if __name__=='__main__':
main()
Following the advice from the dask documentation,
she has to do some manual work first, before she can launch the dask processes.
First, she needs to start the dask-scheduler.
$ dask-scheduler > scheduler.log 2>&1 &
distributed.scheduler - INFO - -----------------------------------------------
distributed.scheduler - INFO - Clear task state
distributed.scheduler - INFO - Scheduler at: tcp://192.168.178.25:8786
distributed.scheduler - INFO - bokeh at: :8787
distributed.scheduler - INFO - Local Directory: /tmp/user/1000/scheduler-k05cez
distributed.scheduler - INFO - -----------------------------------------------
Then, she starts one workers for testing:
$ dask-worker 192.168.178.25:8786 > worker.log 2>&1 &
Lola notices how she has to connect the 2 processes by an IP address
192.168.178.25:8786. After doing all of this, she runs her
script again:
$ python3 distributed.dask_numpi.py
Something has changed. She receives the result much quicker now. Is that
reproducible? Lola measures and observes that the runtime of dask.distributed
is very close to the runtime of the multiprocessing implementation.
By chance, she talks to her office mate about this. They discover that there is
more waiting for them. She opens the URL at 192.168.178.25:8787 in her
browser. Lola is sees an interesting dashboard:
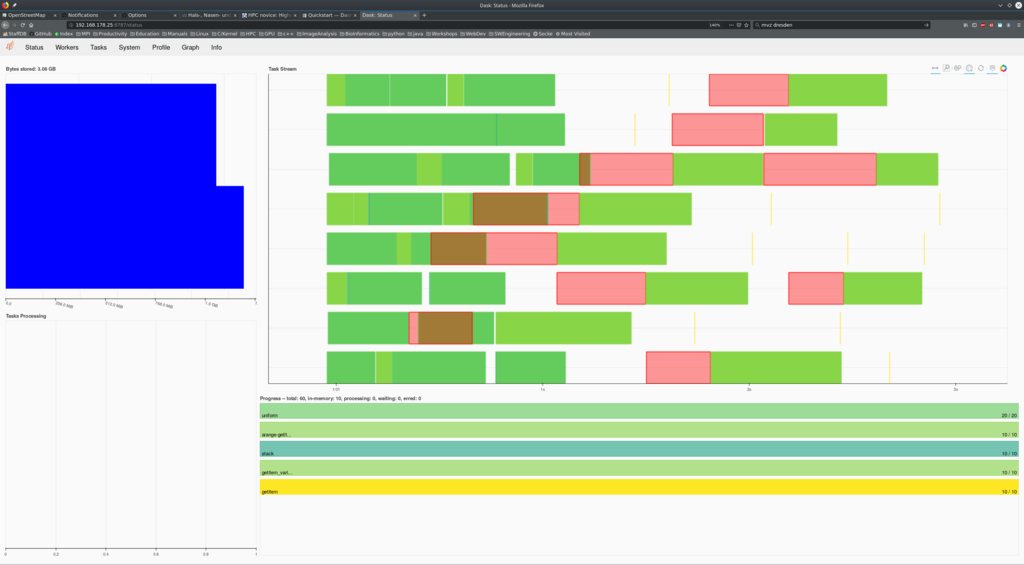
In an cluster environment, this is now a powerful feature. Scaling the
application has just become manageable. So let’s get real and scale across
multiple nodes on the cluster. For this, we start the central dask-scheduler
on the login node. This is a process that only handles network traffic and
hence should not (to be monitored) consume too many resources.
$ dask-scheduler > scheduler.log 2>&1 &
Note, we are sending this process into the background immediately and route its
output including all errors into scheduler.log. The output of this command
should look like this (if not, there is a problem):
distributed.scheduler - INFO - Clear task state
distributed.scheduler - INFO - Scheduler at: tcp://1.1.1.42:8786
distributed.scheduler - INFO - bokeh at: 1.1.1.42:8787
distributed.scheduler - INFO - Local Directory: /tmp/scheduler-xp31e5sl
distributed.scheduler - INFO - -----------------------------------------------
Subsequently, we have to start a worker on a cluster node and connect it to the scheduler by means of the IP address:
$ cat worker.sh
#!/bin/bash
#SBATCH --exclusive
#SBATCH -t 01:00:00
#SBATCH --exclusive
dask-worker tcp://1.1.1.42:8786
$ sbatch -o worker1.log worker.sh
Now, we have to update the address of the scheduler inside our dask python script:
client = Client("tcp://1.1.1.42:8786")
As Lola observes, all parts of this dask system are connected by a single
point, i.e. the IP address of the dask-scheduler. Lola can now run her dask
scripts from the node where the dask-scheduler was started.
$ python3 distributed.dask_numpi.py
She will notice that the dashboard at 1.1.1.42:8787 is now filled with moving
boxes. Her application runs. But, how about adding another node?
$ sbatch -o worker2.log worker.sh
She is curious if the 2 workers can be used by her code.
$ python3 distributed.dask_numpi.py
Lola smiles while looking at the dashboard. This was after all very easy to setup. She has now reached a precision boundary that no other employee has reach for estimating pi.
Key Points
The implementation using multiprocessing used python standard library components (very portable).
The dask library offers parallelisation using constructs that are very numpy like.
To port to dask only the import statements and the container construction needs to be changed.
The advantage of these changes lie in the capability to scale the job to larger machines (test locally, scale globally).
At the heart of the ease of use lie ‘standardized’ building blocks for algorithms using the map-reduce paradigm.
Amdahl’s law still holds.